June 01, 2025
00:00, Sunday, 01 June
May 13, 2025
00:00, Tuesday, 13 May
April 26, 2024
00:00, Friday, 26 April
LLVM Intermediate Representation is an a low-level programming language (more lower than C++, Python etc.).
October 03, 2022
10:00, Monday, 03 October
September 16, 2022
16:00, Friday, 16 September
September 09, 2022
16:00, Friday, 09 September
August 29, 2022
12:35, Monday, 29 August
06:22, Monday, 29 August
August 22, 2022
13:45, Monday, 22 August
August 15, 2022
09:55, Monday, 15 August
July 14, 2022
18:15, Thursday, 14 July
July 08, 2022
18:15, Friday, 08 July
June 29, 2022
18:15, Wednesday, 29 June
April 27, 2022
20:00, Wednesday, 27 April
Thank you to Ondřej Čertík, Oscar Benjamin, Travis Oliphant, and Pamela Chestek for reviewing this blog post. Any errors in the post are my own.
On April 19, 2022, for approximately 11.5 hours, the documentation website for the open source SymPy project and the corresponding GitHub repository were taken down as a result of a DMCA takedown notice submitted by WorthIT Solutions on behalf of HackerRank. In this post, I’d like to explain what the DMCA is and how it works, detail exactly what happened with this incident, and go over my views on the incident and some of the lessons we’ve learned as a result.
What is the DMCA?
The information in this section has been drawn from various sources, including:
- “Your DMCA Safe Harbor Questions Answered” Mitchell Zimmerman (2017)
- The EFF’s Unintended Consequences - 16 Years Under the DMCA (2014)
- GitHub’s DMCA Takedown Policy page
The Digital Millennium Copyright Act (DMCA) is a US law that was passed in 1998. It has laid the groundwork for how copyright is enforced in the age of the internet. One of the DMCA provisions that is important to understand is the so-called “safe harbor” provision. The safe harbor provision, roughly speaking, makes it so that websites can avoid liability because they are simply hosting or transmitting copyrighted content on behalf of their end-users.
The safe harbor provision has been absolutely essential to the modern internet. Without it, a website like GitHub could not exist. Imagine if every time someone found their copyrighted content had been uploaded to GitHub without their permission, that they could sue GitHub for damages. If this were the case, GitHub could never operate. The way GitHub works today, it cannot possibly know if every single thing that is uploaded to it is done so legally. The safe harbor provision also makes modern social media websites like Facebook, Twitter, and YouTube possible.
However, in order for a website to have and maintain safe harbor status, it must follow certain practices as outlined by the DMCA. In particular, sites must manage the “notice-and-take-down process”. GitHub’s notice-and-take-down process is described in its DMCA takedown policy.
The notice-and-take-down process works like this: a provider (like GitHub) has a means for a copyright holder to submit a notice. In GitHub’s case, they provide an online form. This notice must include certain information, including the name and address of the submitter, identification of the copyrighted work, the material on the site that is infringing copyright, and a statement of good faith, signed under penalty of perjury, that the person submitting the notice owns the copyright and believes their rights have been infringed upon. GitHub’s form also requires the submitter to assert that they have taken fair use into consideration.
Once the notice is submitted and once the provider confirms that the above things are done, they must “expeditiously” remove public access to the content. The DMCA does not define “expeditiously”.
The owner of the accused content then has the opportunity to file a counter-notice. GitHub’s counter notice procedure is described in its guide to submitting a DMCA counter notice. This gives the person whose content has been removed recourse if they believe the original notice was invalid. A counter notice contains a statement by the user that the content was removed “as a result of mistake or misidentification”, signed under penalty of perjury. When a provider receives a counter notice, they must forward it to the original complainant, and restore the challenged material after 10-14 business days. At this point, if the complainant wishes to keep the material down, they must file a lawsuit against the alleged infringer.
By following these procedures as outlined by the DMCA, GitHub avoids liability to either party.
GitHub goes beyond what the DMCA law requires. All notices and counter notices that have ever been submitted to GitHub are published publicly in the github/dmca repository (with personal information redacted). GitHub has also historically sided with developers in high profile infringement cases, such as the youtube-dl incident in 2020 (this incident involved a takedown relating to copyright circumvention, which is a different part of the DMCA law that I haven’t discussed here because it isn’t relevant to the notice that was sent to SymPy).
It’s important to understand, however, that GitHub’s hands are tied in many ways here. If they do not follow the notice and counter notice procedures exactly as outlined in the DMCA law, they risk losing their safe harbor status with the US Copyright Office. Were this to happen, it would be completely disastrous to GitHub as a business. Without safe harbor protection, GitHub could be liable for any copyrighted content that someone uploaded to its platform.
Finally, a technical note: when someone submits a takedown notice on a single part of a GitHub repository, GitHub’s response is to take down the entire repository. This is because it is technically impossible to remove only part of a repository due to the way git works. Completely scrubbing data from git history is possible, but it’s a destructive process that’s very disruptive to developers. Instead, GitHub requires the repository owners to scrub the data themselves, if they choose to, within one business day. Otherwise, they will take the entire repository down.
What happened
Below I provide a quick timeline of what happened. Times are in Mountain Daylight Time, which is where I live.
First, for background, the SymPy documentation website https://docs.sympy.org/ is hosted on GitHub Pages and mirrors the repository https://github.com/sympy/sympy_doc. It is common for websites to be hosted on GitHub Pages because it’s completely free and easy to set up for someone who is already used to using git and GitHub. This blog itself is hosted on GitHub Pages. The sympy_doc repository only contains the pre-built HTML of SymPy’s documentation. The actual source code lives in the main SymPy repository. However, the DMCA claims were only ever made against the website, so the main SymPy source code repository was unaffected.
-
Friday, Apr 15, 4:26 PM: Admins of the sympy/sympy_doc GitHub repository (myself, Ondřej Čertík, and Oscar Benjamin) received an automated email from support@githubsupport.com informing us that a DMCA takedown notice was filed against the SymPy documentation. Specifically, the claim, which can now be found on the github/dmca repository, claimed that “Someone has illegally copied our client's technical exams questions and answers from their official website https://www.hackerrank.com/ and uploaded them on their platform without permission.” It specifically referenced the page https://docs.sympy.org/latest/modules/solvers/solvers.html. The notice was made on behalf of HackerRank by an individual from “WorthIT Solutions Pvt. Ltd.” The notice also stated “the infringing website is not willing to remove our client's work”, which came as a surprise to us since, at no point in time prior to receiving this notice had we received any communications from HackerRank or WorthIT Solutions.
The support email stated that if we do not remove the offending content within one business day, the repository, and consequently the docs website, will be taken down.
While it is not relevant to the legality of the situation, it is worth noting that this was on Good Friday and the upcoming Sunday, April 17, was Easter Sunday, which limited our ability to effectively respond over the weekend.
-
Friday, April 15 - Tuesday, April 19: At first, we were not sure if the support email was legitimate or if it was just convincing spam. One thing that confused us was that the support email came from a domain, githubsupport.com, which did not appear to exist. Another issue is that although GitHub’s policy was to post all DMCA claims to the github/dmca repository, this particular claim had not yet been uploaded there. We reached out to GitHub support to ask if the email was legitimate, and they responded Monday, April 18, 3:18 PM that the email was indeed a legitimate email from GitHub Trust & Safety.
I had already reached out on Friday to NumFOCUS for assistance. NumFOCUS is a 501(c)(3) non-profit organization that represents many open source scientific projects including SymPy. However, due to the limited time frame of the notice and the fact that it occurred over a holiday weekend, NumFOCUS was not able to connect us with legal counsel until Tuesday.
-
Tuesday, April 19, 12:21 PM: We received notice from GitHub that the sympy/sympy_doc repository has been taken offline. The sympy/sympy_doc repository was replaced with a notice that the documentation was taken down by the DMCA notice with a link to the notice, which had been posted to the public github/dmca repository, and the documentation website itself started to 404. Since this was now public knowledge, we publicly announced this on the SymPy mailing list and Twitter.
We also worked with NumFOCUS and the legal counsel to send a counter notice to GitHub, since we believed that the notice was mistaken and that the claimed infringement, the code and mathematical examples on the docs page, were likely not even copyrightable.
Not long after the takedown occurred, someone posted it to Hacker News. The Hacker News posting eventually made its way to the front page and by the end of the day it had made its way to rank 3 on the site, where it received hundreds of upvotes and comments. The story also generated a lot of buzz on Twitter at this time.
-
Tuesday, April 19, 6:40 PM: Vivek Ravisankar, the CEO of HackerRank, posted a public apology on the Hacker News thread:
Hello, I'm Vivek, founder/CEO of HackerRank. Our intention with this initiative is to takedown plagiarized code snippets or solutions to company assessments. This was definitely an unintended consequence. We are looking into it ASAP and also going to do an RCA to ensure this doesn't happen again. Sorry, everyone! -
Tuesday, April 19, 8:26 PM: Vivek posted a followup comment on Hacker News:
Hello again, Vivek, founder/CEO here. In the interest of moving swiftly, here are the actions we are going to take: (1) We have withdrawn the DMCA notice for sympy; Sent a note to senior leadership in Github to act on this quickly. (2) We have stopped the whole DMCA process for now and working on internal guidelines of what constitutes a real violation so that these kind of incidents don't happen. We are going to do this in-house (3) We are going to donate $25k to the sympy project. As a company we take a lot of pride in helping developers and it sucks to see this. I'm extremely sorry for what happened here.HackerRank did follow through with the $25k donation to SymPy.
-
Wednesday, April 20, 12:00 AM (approximately): The SymPy documentation website and the sympy/sympy_doc repository went back online.
-
Wednesday, April 20, 2022, 10:11 AM: We received an email notice from GitHub that the DMCA claim against our repository has been retracted. The retraction is made available online on the github/dmca repository.
My Views
Now that I’ve stated the facts, let me take a moment to give my own thoughts on this whole thing. My belief is that the DMCA claim that was made against the SymPy repository was completely baseless and without merit. Not only is no part of the SymPy documentation, to my knowledge, taken illegally from HackerRank or any other copyrighted source, but the claim itself was completely unspecific about which parts of the documentation were supposedly infringing. This made it impossible for us to comply even if the complaint was legitimate. In addition, it is questionable whether the supposed parts of the documentation that were taken from HackerRank, the mathematical and code examples, are even copyrightable. While this may not have been an actively targeted attack against SymPy, as a community run open source project, it served as one in practice.
We have never learned any more about which parts of the SymPy documentation were supposedly infringing on HackerRank’s copyright, and I expect we never will. Some people online have speculated that HackerRank actually took examples from the SymPy documentation, not the other way around, but I do not know if this is the case or not. It seems likely, given the large number of other takedown notices WorthIT has made to GitHub on HackerRank’s behalf, that they were using some sort of automated or semi-automated detection system, which somehow detected what it thought was infringing content on our website. I have personally never used the HackerRank platform, so I only know what sorts of things are on it from second-hand accounts.
Firstly let me say that if anyone or anything is to be blamed for this, my personal belief is that the blame should primarily lie on the DMCA law itself. While the “safe harbor” provisions of the law have been critical to the development of the modern internet, allowing social websites such as GitHub to exist without the burden of legal liability for user contributed content, other provisions are hostile to those same users, even those who are operating in a completely legal manner. The DMCA works in a “guilty until proven innocent” way, and the very operation of the takedown notice policy implicitly assumes that those making claims are not abusing the system as bad actors. It also incentivizes platforms such as GitHub to step aside and not take sides in claims, even claims such as this one, which refer to likely uncopyrightable material or are too unspecific to reasonably address even if they are legitimate.
The DMCA also has had further chilling effects due to its anti-circumvention provisions. The EFF has written extensively about these. While something as radical as abolishing copyright may be considered to be practically untenable, I believe that laws like the DMCA can be rewritten so that they still serve their intended function of protecting copyright owners and providing “safe harbor” to websites like GitHub, while also protecting the rights of those accused of infringing copyright.
With that being said, I think some blame does need to lie on HackerRank and WorthIT Solutions for abusing the DMCA system and filing claims like this. They also lied in their claim when they said “the infringing website is not willing to remove our client's work.” At no point were we contacted by HackerRank or WorthIT Solutions about any copyright infringement. If we were, we would have been happy to work with them to determine whether any content in SymPy is infringing on copyright and remove it if it was. Even if it were the case that SymPy’s documentation infringed on HackerRank’s copyright, the immediate escalation to a DMCA takedown notice, which legally forced GitHub to completely disable the entire SymPy documentation, was completely inappropriate. Even submitting a counter notice is risky, because by doing so it increases the likelihood that the complaining party will bring an infringement lawsuit. Many DMCA’d repositories do not submit a counter notice for this reason, and we were only able to do so comfortably because we were able to do so via NumFOCUS. Had HackerRank not retracted the notice, we would have had to wait for our counter notice to take effect, meaning our docs website would have remained unavailable for 10-14 days. And had they instead taken down the main SymPy repository instead of the sympy_doc repo where the docs are hosted, this would have been a major disruption for our entire development workflow.
I do want to thank Vivek for quickly retracting the notice once this came to his attention, and I hope that he will be rethinking their DMCA policies at HackerRank, and for their donation to support SymPy and NumFOCUS.
Finally, while many have blamed GitHub, I believe that for the most part, they have acted as they are effectively required to by law. It is important to understand that any similar website based in the US would have likely treated this claim in a similar way. For instance, here is GitLab’s DMCA policy, which you can see is very similar to GitHub’s. By all accounts, GitHub is an industry leader here, by posting all notices and counter notices online, which they are not required to do, and by being very open about their DMCA policies ([1], [2], [3]). With that being said, there are ways that GitHub’s processes could be improved here. The one business day that we were given to respond is actually standard in the industry, and ultimately comes from the “expeditiously” wording of the DMCA. But even so, if we were given more time than this, it would have made things much easier. It may have even been possible for us to reach out to HackerRank and resolve the situation before any actual takedown occurred. It also would be helpful if GitHub, while staying within the DMCA provisions, had a higher standard of legitimacy for takedown notices such as the one we received. As I have noted, the request we received was not nearly specific enough for us to effectively act on it, which should have been apparent to GitHub, since the only information WorthIT provided was a link to the HackerRank home page. Finally, I have two small technical requests from GitHub: first is to make githubsupport.com a real website so that support emails appear more legitimate, and second is to post DMCA notices to the DMCA repository right away rather than only after the repository is taken down, to make it clearer that the notices are in fact legitimate.
Open source communities such as SymPy are under-resourced and typically ill equipped to handle situations like these, which inherently puts them in an inferior position. This is despite the fact that open source software serves a global commons that benefits all. I have estimated that SymPy itself is used by hundreds of thousands of people, and the broader scientific Python ecosystem that it is part of is used by millions of people. Abuses of copyright law against these communities are harmful to society as a whole.
Lessons Learned
I’d like to end with a few lessons that I’ve learned from this whole process.
- Take DMCA takedown notices seriously. If you ever receive a DMCA takedown notice from GitHub or any other website, you need to take it seriously. The website is required by law to remove your content after they receive such a notice. You can submit a counter notice, but this increases the likelihood of being sued.
- NumFOCUS fiscal sponsorship is invaluable. Our NumFOCUS fiscal sponsorship was invaluable here. Thanks to NumFOCUS, we were able to immediately access high quality legal advice on how to proceed here. Had HackerRank not retracted their notice, or even, heaven forbid, decided to sue, we would have had significant assistance and support from NumFOCUS. If you are part of an open source project that doesn’t have fiscal sponsorship, I would recommend looking into NumFOCUS, or other similar organizations. And if you want to support NumFOCUS, I encourage you to do so.
- Make backups of your online data. While the source code of any GitHub repository is effectively backed up onto every computer that has a git clone. Other data such as issues and pull request comments are not. We were lucky that the DMCA notice was sent against our documentation repository instead of our main repository. If our main repository were taken down instead, we would have lost access to all GitHub issue and pull request history. I have been looking into effective ways to backup this data. If anyone has any suggestions here, please let me know in the comments.
I do feel that being on a site like GitHub is still preferable to something like self-hosting. If you self-host, you become responsible for a ton of things which GitHub does for you, like making sure everything stays online, handling servers, and managing spam. Also, self-hosting does not magically shield you from legal threats. If you self-host content that infringes on someone’s copyright, you are still legally liable for hosting that content.
Finally, I want to thank everyone who supported SymPy during this incident. Even if you just talked about this on social media or upvoted the Hacker News story, that helped us get this into the public eye, which led to a much faster resolution than we expected. I especially want to thank Leah Silen and Arliss Collins from NumFOCUS; Ondřej Čertík, Oscar Benjamin, and other SymPy community members; Pamela Chestek, the NumFOCUS legal counsel; and Travis Oliphant for the help they provided to us. Thank you to Thomas Dohmke, GitHub’s CEO, who we have reached out to privately, and who has promised to improve GitHub’s DMCA policies. I also want to thank Vivek Ravisankar from HackerRank for retracting the DMCA claim, and for the generous donation to SymPy.
Now that this incident is over, I’m hopeful we in the SymPy community can all go back to building software.
If you wish to donate to support SymPy, you may do so here.
July 12, 2021
11:45, Monday, 12 July
|
|
|
Hi there! It’s been five weeks into GSoC & I have managed to refactor remaining 3 solvers. With this, I completed all the work which I proposed for my GSoC project. Although I will be working further to make some enhancements in terms of improving speed of dsolve.
The solvers which I managed to refactor are:
- 2nd_linear_airy: It gives solution of the Airy differential equation

in terms of Airy special functions airyai and airybi.
During refactoring the matching code was also simplified as earlier it used to match the equation and preprocess the coefficients accordingly. But now internal function named get_linear_coeffs is used to extract the coefficients and then try to match with the solver.
>>> from sympy import dsolve, Function
>>> from sympy.abc import x
>>> f = Function("f")>>> eq = f(x).diff(x, 2) - x*f(x)
>>> dsolve(eq, hint = '2nd_linear_airy')
Eq(f(x), C1*airyai(x) + C2*airybi(x))
- 2nd_linear_bessel: It gives solution of the Bessel differential equation

if n is integer then the solution is of the form Eq(f(x), C0 besselj(n,x) + C1 bessely(n,x)) as both the solutions are linearly independent else if n is a fraction then the solution is of the form Eq(f(x), C0 besselj(n,x) + C1 besselj(-n,x)) which can also transform into Eq(f(x), C0 besselj(n,x) + C1 bessely(n,x)).
>>> from sympy.abc import x
>>> from sympy import Symbol
>>> v = Symbol('v', positive=True)>>> from sympy.solvers.ode import dsolve
>>> from sympy import Function
>>> f = Function('f')>>> y = f(x)
>>> genform = x**2*y.diff(x, 2) + x*y.diff(x) + (x**2 - v**2)*y
>>> dsolve(genform)
Eq(f(x), C1*besselj(v, x) + C2*bessely(v, x))
Here is the Patch for above 2 solvers.(Merged)
- lie_group: This hint implements the Lie group method of solving first order differential equations. The aim is to convert the given differential equation from the given coordinate system into another coordinate system where it becomes invariant under the one-parameter Lie group of translations. The converted ODE can be easily solved by quadrature.
In sympy lie_group solver had several heuristics functions implemented which were kept in dsolve.py, So all the helper functions were moved to separate file and a wrapper class was implemented to match and return the solutions.
>>> from sympy import Function, dsolve, exp, pprint
>>> from sympy.abc import x
>>> f = Function('f')>>> pprint(dsolve(f(x).diff(x) + 2*x*f(x) - x*exp(-x**2), f(x),
... hint='lie_group'))
/ 2\ 2
| x | -x
f(x) = |C1 + --|*e
\ 2 /
Here is the Patch for lie_group.
That’s all for now, looking forward for week #6.
July 03, 2021
17:10, Saturday, 03 July
|
|
|
Hi there! It’s been four weeks into GSoC & I have managed to refactor six more solvers. Week 3 included lot of brainstorming around the simplification of the solutions returned by the solvers.
The solvers which I managed to refactor are:
- nth_linear_constant_coeff_homogeneous: Solves an nth order linear homogeneous differential equation with constant coefficients.This is an equation of the form :

These equations can be solved in a general manner, by taking the roots of the characteristic equation

>>> from sympy import Function, dsolve, pprint
>>> from sympy.abc import x
>>> f = Function('f')>>> pprint(dsolve(f(x).diff(x, 4) + 2*f(x).diff(x, 3) -
... 2*f(x).diff(x, 2) - 6*f(x).diff(x) + 5*f(x), f(x),
... hint='nth_linear_constant_coeff_homogeneous'))
x -2*x
f(x) = (C1 + C2*x)*e + (C3*sin(x) + C4*cos(x))*e
- nth_linear_constant_coeff_variation_of_parameters: Solves an nth order linear differential equation with constant coefficients using the method of variation of parameters.This method works on any differential equations of the form

This method works by assuming that the particular solution takes the form

where y_i is the ith solution to the homogeneous equation. The solution is then solved using Wronskian’s and Cramer’s Rule. The particular solution is given by

where W(x) is the Wronskian of the fundamental system (the system of n linearly independent solutions to the homogeneous equation), and W_i(x) is the Wronskian of the fundamental system with the ith column replaced with [0, 0, …., 0, P(x)].
- nth_linear_constant_coeff_undetermined_coefficients: Solves an nth order linear differential equation with constant coefficients using the method of undetermined coefficients. This method works on differential equations of the form

where P(x) is a function that has a finite number of linearly independent derivatives.
# Generating trialset for P(x)
>>> from sympy import log, exp, Function
>>> from sympy.solvers.ode.single import NthLinearConstantCoeffUndeterminedCoefficients,SingleODEProblem
>>> from sympy.abc import x
>>> f = Function('f')>>> eq = 9*x*exp(x) + exp(-x)
>>> obj = NthLinearConstantCoeffUndeterminedCoefficients(SingleODEProblem(eq,f(x),x))
>>> obj._undetermined_coefficients_match(eq,x)
{'test': True, 'trialset': {x*exp(x), exp(-x), exp(x)}}Note : Both nth_linear_constant_coeff_undetermined_coefficients and nth_linear_constant_coeff_variation_of_parameters require roots of the characterstic equations so instead of inheriting nth_linear_constant_coeff_homogeneous they create an instance of it and reuse the solutions method.
Here is the Patch for above 3 solvers.(Merged)
- nth_linear_euler_eq_homogeneous: Solves an `n`\th order linear homogeneous variable-coefficient Cauchy-Euler equidimensional ordinary differential equation. This is an equation with form

These equations can be solved in a general manner, by substituting solutions of the form f(x) = x^r , and deriving a characteristic equation for r.
>>> from sympy import Function, dsolve, pprint
>>> from sympy.abc import x
>>> f = Function('f')>>> eq = f(x).diff(x, 2)*x**2 - 4*f(x).diff(x)*x + 6*f(x)
>>> pprint(dsolve(eq, f(x),
... hint='nth_linear_euler_eq_homogeneous'))
2
f(x) = x *(C1 + C2*x)
- nth_linear_euler_eq_nonhomogeneous_variation_of_parameters: Solves an `n`\th order linear non homogeneous Cauchy-Euler equidimensional ordinary differential equation using variation of parameters.This is an equation with form

Rest of the procedure is same as nth_linear_constant_coeff_variation_of_parameters.
- nth_linear_euler_eq_nonhomogenus_undetermined_coefficients: This is an equation with form

These equations can be solved in a general manner, by substituting solutions of the form f(x) = x^r and then deriving a characteristic equation for r and rest of the process is same as nth_linear_constant_coeff_undetermined_coefficients.
Here is the patch for the euler solvers.
That’s all for now, looking forward for week #5.
June 20, 2021
11:05, Sunday, 20 June
|
|
|
Hi there! It’s been two weeks into GSoC & I have managed to refactor two more solvers.Because of my university exams, I started this week’s work from wednesday.
This week the solvers which I managed to refactor are:
- nth_order_reducible: For example any second order ODE of the form f’’(x) = h(f’(x), x) can be transformed into a pair of 1st order ODEs g’(x) = h(g(x), x) and f’(x) = g(x). Usually the 1st order ODE for g is easier to solve. If that gives an explicit solution for g then f is found simply by integration.
The patch can be found here.(Merged) - 2nd_hypergeometric: It computes special function solutions which can be expressed using the 2F1, 1F1 or 0F1 hypergeometric functions.
y’’ + A(x) y’ + B(x) y = 0 where `A` and `B` are rational functions.
These kinds of differential equations have solution of non-Liouvillian form. Given linear ODE can be obtained from 2F1 given by
(x² — x) y’’ + ((a + b + 1) x — c) y’ + b a y = 0 where {a, b, c} are arbitrary constants.
Currently only the 2F1 case is implemented in SymPy.
The patch can be found here. (Documentation was also added for this solver 📝)
That’s all for now, looking forward for week #3.
June 12, 2021
14:33, Saturday, 12 June
|
|
|

Google Summer of Code is a global program focused on bringing more student developers into open source software development. Students work with an open source organization on a 10 week programming project during their break from school.
What is SymPy?

SymPy is a Python library for symbolic mathematics. It aims to become a full-featured computer algebra system (CAS) while keeping the code as simple as possible in order to be comprehensible and easily extensible.
I have been contributing to Sympy organisation for more than a year now. Last year, I applied for the same but couldn’t get through. But this was not the case this time.On 17th May, results were announced and I got to know that my project was selected.
https://medium.com/media/0928bd7a31ead3ca386cb9beaade863b/hrefAbout my Project
My project is being mentored by Aaron meurer and Oscar benjamin.
The project aims at Refactoring of ODE module. Currently, the dsolve function in the ODE module is a bit messy, as whenever dsolve is called for solving an ODE, it first calls classify_ode() which tries to match each solver and after that, it again calls that particular solver for solving. If a particular solver matches the equation it should directly return the solution instead. So, sometimes the solver which returns the solution is much faster than running all the matches.
My detailed proposal for the project can be found here.
Progress of Week I and Community bonding period
Since I have been contributing to the SymPy for a good amount of time, it was easier for me to get into the community. So, in this period I discussed implementation details of the workflow with my mentors and we concluded that initially each solver should be moved to classes with generalised methods and their associations.
In community bonding period I refactored three solvers:
- Liouville solver : It solves 2nd order Liouville differential equations.
General form for which it works : d²y/dx² + g(y)*(dy/dx)² + h(x)*dy/dx.
Here is my patch for the same. (Merged) - Separable solver : It solves 1st order differential equations.
General form for which it works : P(y)dy/dx=Q(x).
Here is my patch for the same. (Merged) - Separable Reduced solver : It solves 1st order differential equations and
converts it into separable equation.
General form for which it works : y′+(y/x) H((x^n) y)=0.
Here is my patch for the same. (Merged)
During my first week of coding period , I managed to refactor four solvers:
- 1st_homogeneous_coeff_subs_dep_div_indep : It works on homogenous ODE of the form P(x, y) + Q(x, y) dy/dx = 0 such that P and Q are homogeneous and of the same order. Here the substitution will be <dependent variable>/<independent variable>.
- 1st_homogeneous_coeff_subs_indep_div_dep : It works on homogenous ODE of the form P(x, y) + Q(x, y) dy/dx = 0 such that P and Q are homogeneous and of the same order. Here the substitution will be <independent variable>/<dependent variable>.
- 1st_homogeneous_coeff_best : It just calls the solution from above two methods and return the best simplified solution among them.
- Linear_coefficients : General form of this type:
y′+F((ax+by+c)/(dx+ey+f))=0. This can be solved by substituting
x=x′+(e*c−b*f)/(d*b−a*e) and y=y′+(a*f−d*c)/(d*b−a*e). This substitution reduces the equation to a homogeneous differential equation.
As all four solvers were interdependent used to call each other I made a single PR for them. It can be found here. (Merged)
That’s all for now, looking forward for week #2.
June 03, 2021
03:57, Thursday, 03 June
I've ditched Disqus as the comment system on this blog. I am now using Utterances. Utterances is an open source comment system that is backed by GitHub issues. Basically, every post has a corresponding issue opened on GitHub, and the comments on the post are the comments on the issue. Utterances automatically places the comments at the bottom of the post. For example, here is the issue corresponding to this post.
I didn't like Disqus mostly because it serves ads and tracking. Even though I had opted out from as much of it as I could in the Disqus settings, it still loads tracking scripts on every page. I run uBlock Origin always, and it's a bit hypocritical if my own side has things that are blocked by it. In some cases I can't avoid it (as far as I know), like when I embed a YouTube video, but it definitely shouldn't be on every post.
Utterances is a very nice alternative. I has lots of advantages:
- Comments are not self-hosted. GitHub hosts them. Since you need a GitHub account to comment, this should make comment spam a non-issue.
- Comments support full Markdown.
- Users can edit their comments.
- I can edit and fully moderate all comments.
- Users log in with a federated system that proves their identity.
- Email subscription to posts.
- No ads or tracking.
- It's completely open source and free to use.
If you use Nikola like I do, it natively supports Utterances (a feature which I added). Otherwise, go to the Utterances and paste the script tag generated at the bottom into your blog template. Then install the Utterances app in your repo, and you are done.
Exporting Disqus Comments
Some of my old posts had Disqus comments, which I wanted to preserve somehow. Here is guide on how I did that, since it wasn't as straightforward as I would have hoped.
The first step is to export your Disqus comments. It's very difficult to actually find the place in the Disqus site where you do this, but I finally found the URL. The export takes some time to complete (for me it took about half an hour). When it finished, Disqus will email you an XML file with all your comments. Note that the file contains all comments for all sites you have ever set up with Disqus. For me, it also included all the comments on my old Wordpress blog, as well as posts for draft blog posts that I never ended up publishing. It also contained all comments that were marked as spam, so you will need to remember to filter those.
I decided that since I only have a handful of posts with Disqus comments, I would just write a script to process them all and manually print them out, which I will then manually enter in to the Utterances comment system for those posts.
I wrote a script to process the comments, which you can find here. Disqus does provides an XML schema for the XML. I used a library called xsData, which lets you take an XML scheme and generate Python dataclasses corresponding to it, which make manipulating the parsed XML much easier than the standard library xml library. The script outputs text like
========== Comments from https://asmeurer.github.io/blog/posts/what-happens-when-you-mess-with-hashing-in-python/ ==========
These are the original comments on this post that were made when this blog used the [Disqus blog system](https://www.asmeurer.com/blog/posts/switching-to-utterances-comments/).
>**Comment from bjd2385 on 2016-08-28 12:33:12+00:00:**
><p>Very interesting post. I was just looking into hash functions (I've never formally learned what the topic entails), and since I'm most familiar with Python this post explained quite a bit, especially your early mathematical points.</p>
>**Comment from Mark Lawrence on 2016-10-03 20:26:54+00:00:**
><p>At what point does Python 3 force the override of __hash__ if you've defined __eq__? E.g when would your</p><p>AlwaysEqual class fail?</p>
>**Replies:**
>>**Comment from asmeurer on 2016-10-03 20:38:13+00:00:**
>><p>That's a good catch. I originally wrote this post in Python 2. The example does indeed fail in Python 3. More specifically, if you override __eq__, Python 3 automatically sets __hash__ to None. I'll update the post to make this more clear.</p>
>**Comment from Erick Mendonça on 2017-07-30 03:23:55+00:00:**
><p>Great article! We must really pay attention to these details when implementing custom hashes.</p>
>**Comment from Ignacio on 2017-10-07 22:31:56+00:00:**
><p>Thanks a lot for this post! Clarified a lot of concepts.</p>
which I then manually copied to each post's Utterances page on GitHub.
Feel free to adapt my script if you find yourself in a similar situation.
Utterances Comments
Feel free to use the comments on this page to play around with the commenting system.
Note that to comment, there are two options. You can log in on this page, which will let you type your comment in the box below. This requires giving the Utterances bot access to your GitHub account. Alternately, if you don't want to give a bot access, you can just go directly to the GitHub issue page and comment there. I am currently in the process of figuring out how to add some boilerplate to each page that makes this clear (see this Utterances issue). If anyone has any suggestions on how to do this, let me know. For now, I am just going to manually add a statement about this as the first comment on each post.
October 30, 2020
18:30, Friday, 30 October
This summer I had plenty of time during COVID-19 lockdown and I was looking at SymPy Gamma.
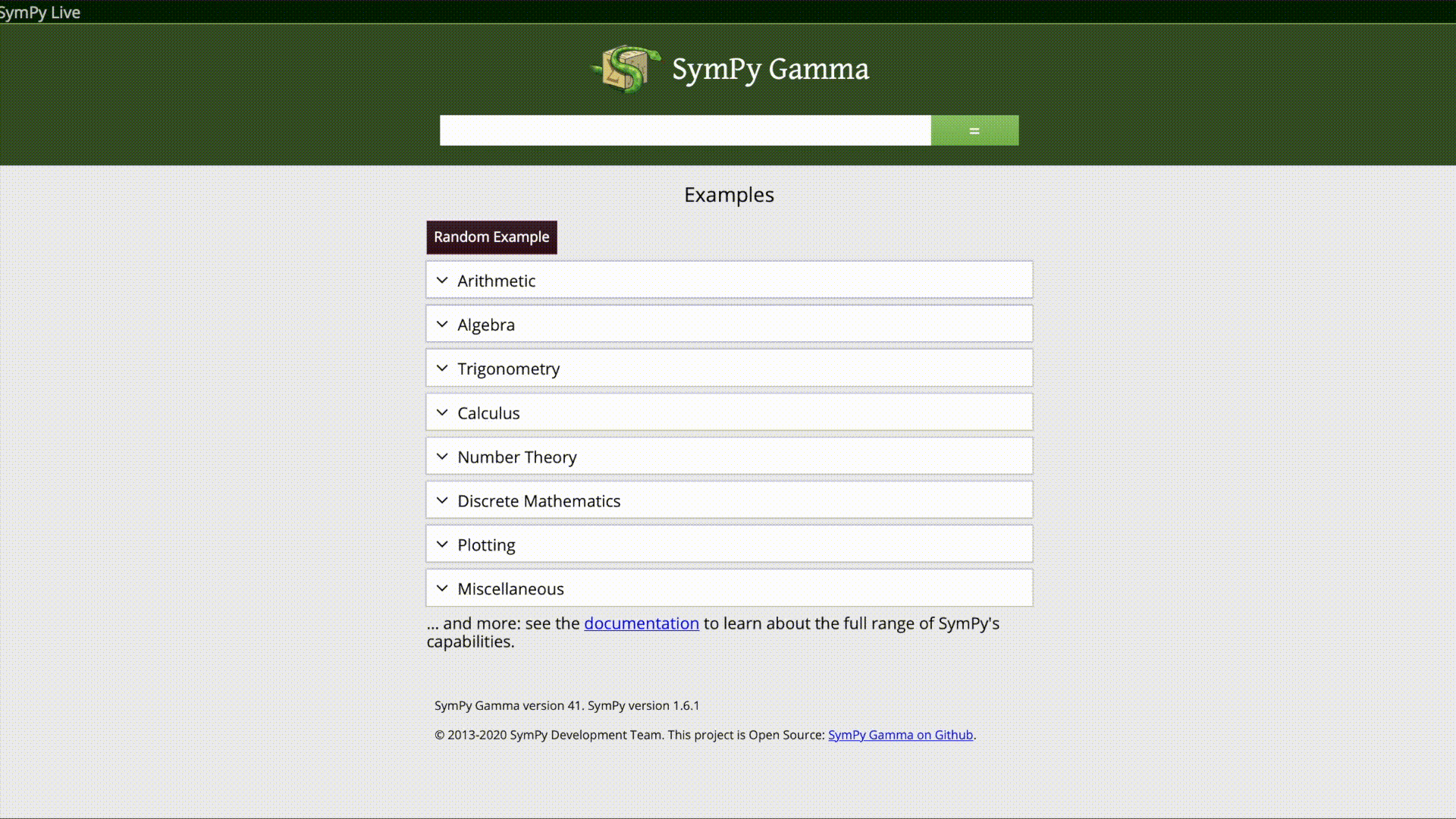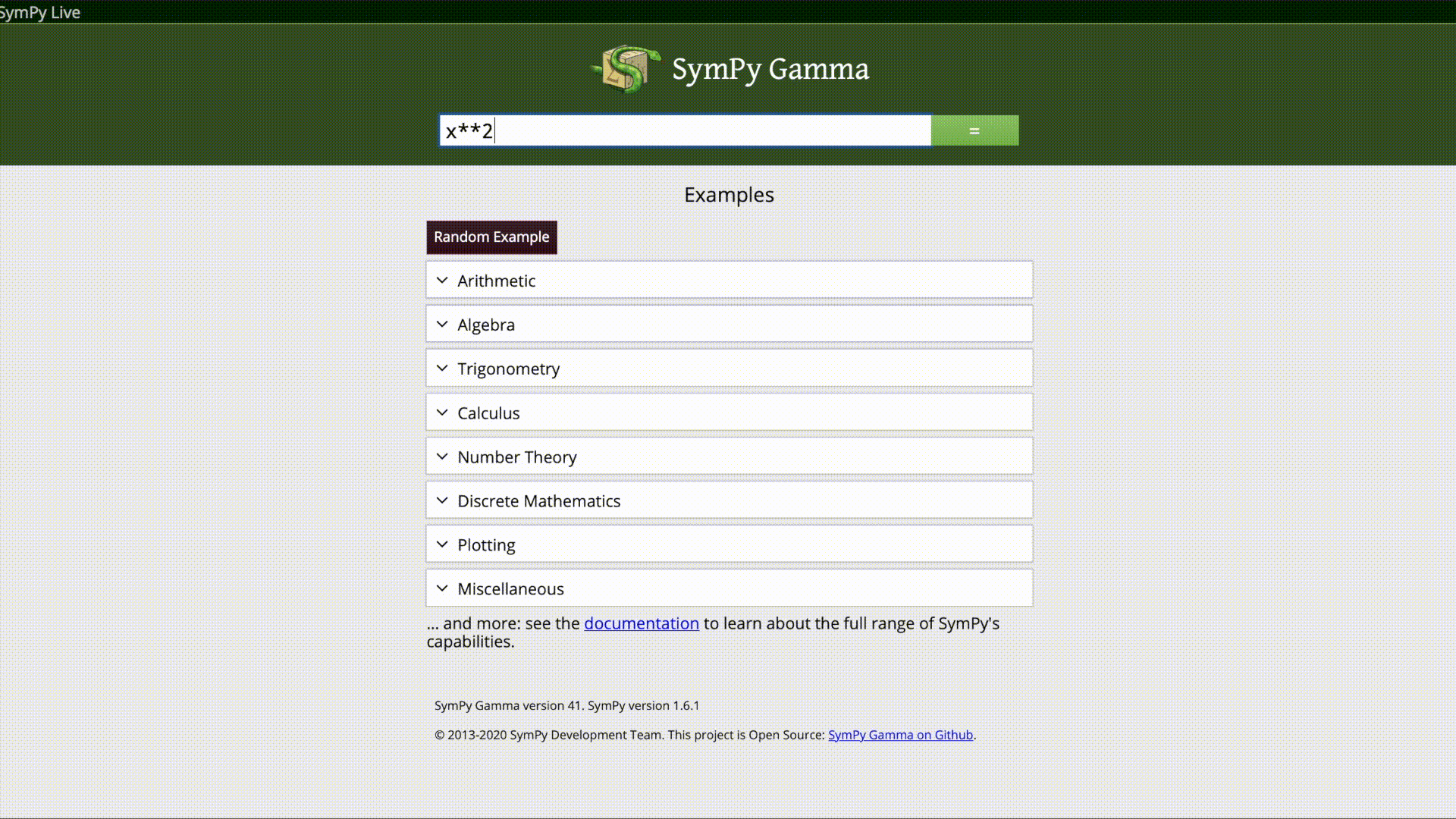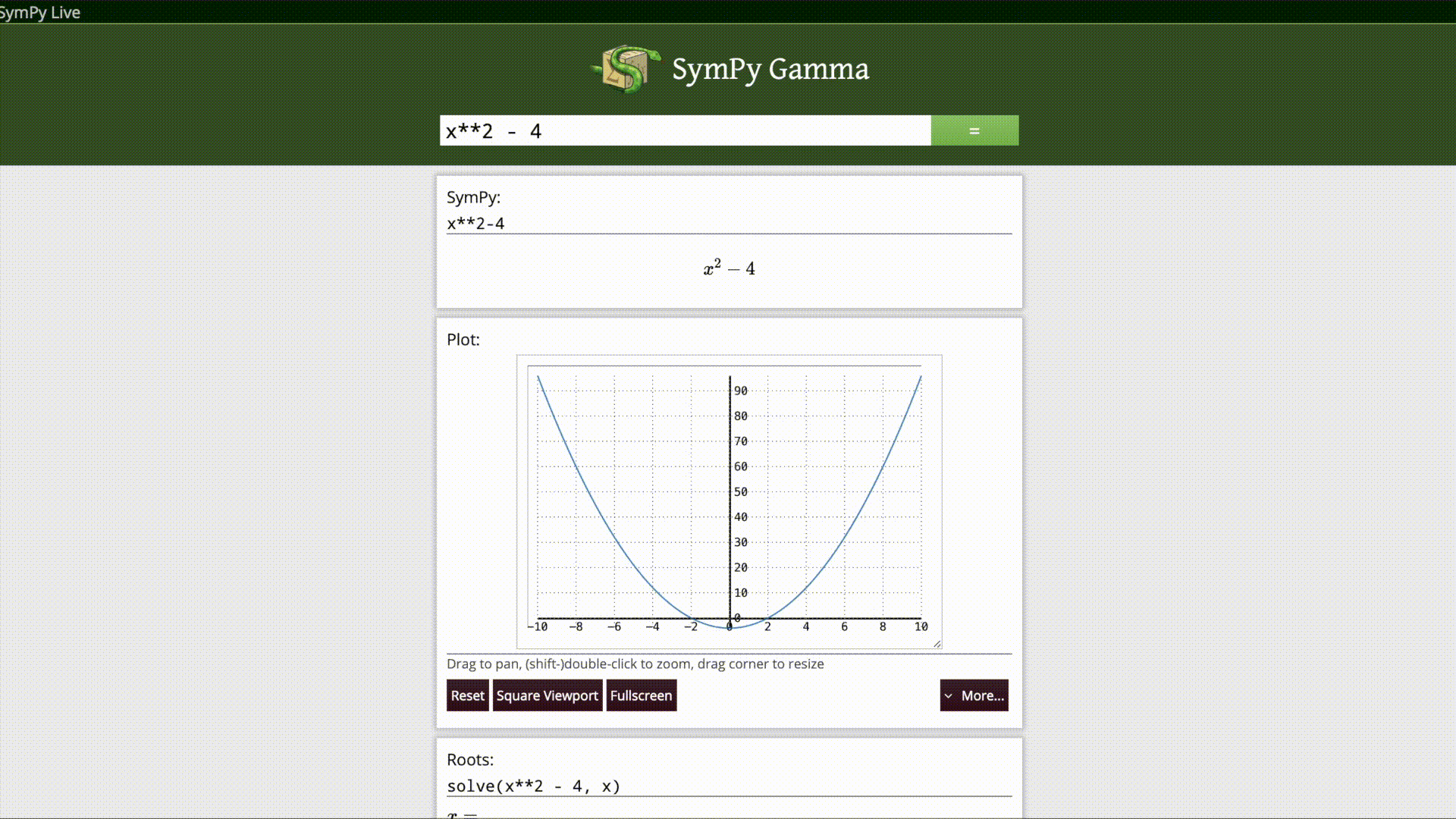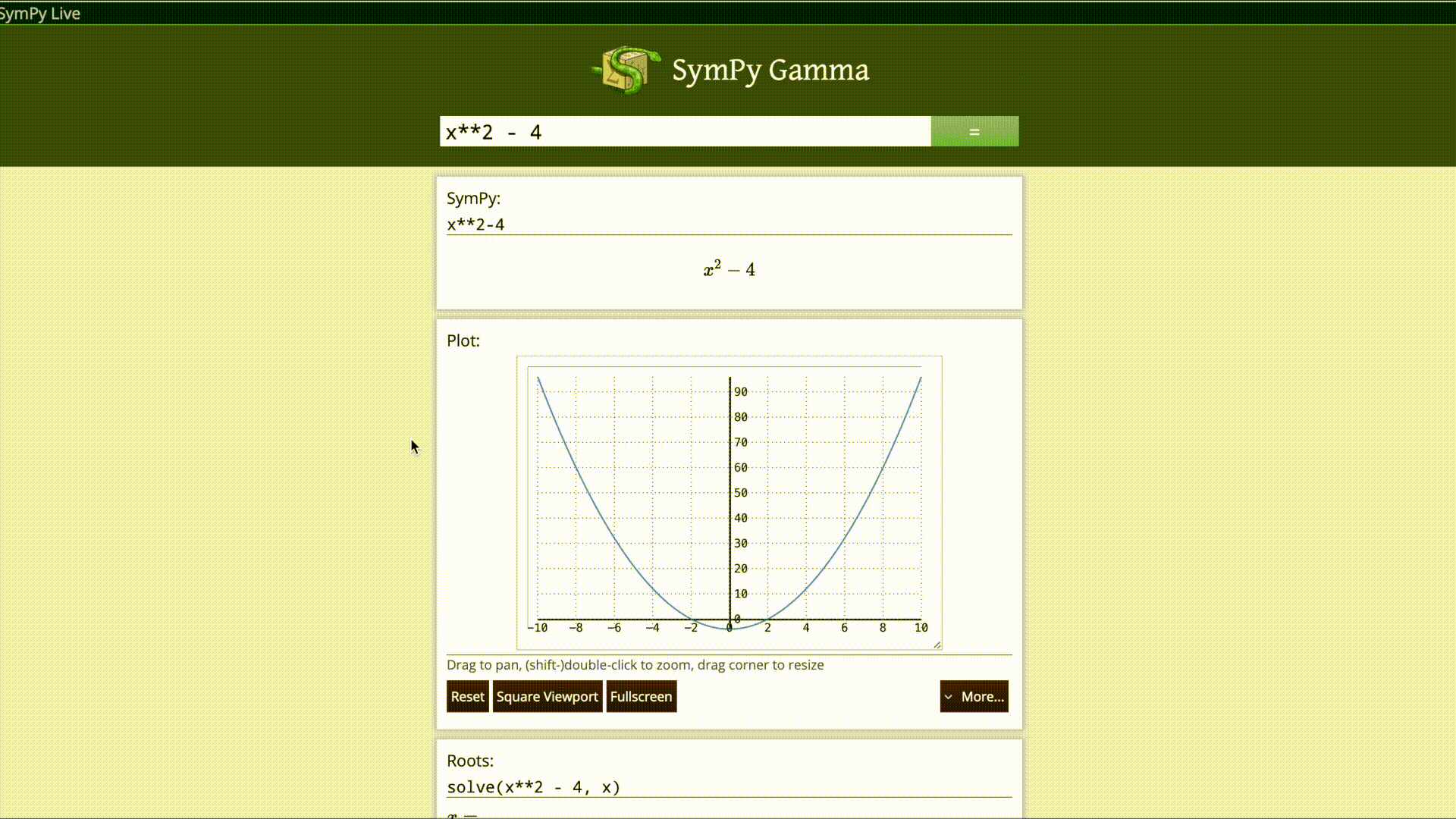
Sympy Gamma
SymPy Gamma is a web application that executes mathematical expressions via natural language input from the user, after parsing them as SymPy expressions it displays the result with additional related computations. It is inspired from the idea of WolframAlpha which is based on the commercial Computer Algebra System named “Mathematica”.
I have always been impressed by it ever since I first found about it. While playing with it during this summer, I realised that it runs on Google App Engine’s Python 2.7 runtime. It is powered by SymPy, an open source computer algebra system.
The Problem
Despite being widely used around the world (about ~14K users everyday, as seen from Google Analytics), there hasn’t been a lot of development in the past 5 years. Due to this the current infrastructure was stuck on Google App Engine’s Python 2 runtime which obviously does not support Python 3.
This also prevented it to use the latest version of SymPy. The SymPy version (~0.7.6) it was using was last updated 6 years ago. This made SymPy Gamma in urgent need for upgradation. At the time of writing this blog, SymPy Gamma is running on Google App Engine’s latest runtime and latest version of SymPy.
Solution and Process
It was a fun project and was interesting to see how Google cloud offering has evolved from Google App Engine to Google Cloud Platform. The old App engine did seem like a minimal cloud offering launched by Google in an attempt to ship something early and quickly. It reminded me of my first cloud project in college (dturmscrap), which I deployed to Google App Engine, back in 2015.
I used Github projects to track the whole project, all the work done for this can be seen here.
$ Git Log
Here is a summary of what was achieved:
-
PR 135: Migrating Django to a slightly higher version, this was the first blood just to make sure everything was working. I upgraded it to the latest version of Django that was supported on Python 2 runtime. This also exposed the broken CI, which was fixed in this.
-
PR 137: This upgraded the CI infrastructure to use Google Cloud SDK for deployment, the previous method was discontinued.
-
PR 140: Upgrading the Database backend to use Cloud NDB instead of the legacy App Engine NDB.
-
PR 148: Since we change the database backend, we needed something for testing locally, this was done by using Google Cloud Datastore emulator locally.
-
PR 149: The installation and setup of the project was quite a challenge. Installing and keeping track of the versions of a number of application was non-trivial. This Pull request dockerized the project and made the development setup trivial and it all boiled down to just one command.
-
PR 152: The login feature was previously implemented using the user API of the Google App Engine’s Python2 runtime, which was not available in Python 3 runtime. We removed the login feature as it was not used by many and was too much effort to setup OAuth for login.
-
PR 153: Now was the time to slowly move towards Python 3 by making the code compatible with both 2 and 3. It was achieved via python-modernize.
-
PR 154: We then made the migration to Python 3.7 runtime and removed submodules and introduced a
requirements.txtfor installing dependencies. -
PR 159: The above change made it possible to upgrade SymPy to latest version, which was 1.6 at that time.
-
PR 165: The last piece of the puzzle was upgrading Django itself, so we upgraded it to the latest version, which was Django 3.0.8.
Next Steps
- At the time of writing this Google has released the Python 3.8 runtime, it would nice to further upgrade it now.
- The test coverage can be increased.
- The code can be refactored to be more readable and approachable for new contributors.
Thanks to Google for properly documenting the process, which made the transition much easier.
Thanks to NumFocus, without them this project would not have been possible. Also thanks to Ondrej Certik and Aaron Meurer for their advice and support throughout the project.
August 31, 2020
00:00, Monday, 31 August
This report summarizes the work done in my GSoC 2020 project, Implementation of Vector Integration with SymPy. Blog posts with step by step development of the project are available at friyaz.github.io.
About Me
I am Faisal Riyaz and I have completed my third year of B.Tech in Computer Engineering student from Aligarh Muslim University.
Overview
The goal of the project is to add functions and class structure to support the integration of scalar and vector fields over curves, surfaces, and volumes. My mentors were Francesco Bonazzi (Primary) and Divyanshu Thakur.
Work Completed
Here is a list of PRs which were opened during the span of GSoC:
-
(Merged) #19472: Adds
ParametricRegionclass to represent parametrically defined regions. -
(Merged) #19539: Adds
ParametricIntegralclass to represent integral of scalar or vector field over a parametrically defined region. -
(Merged) #19580: Modified API of
ParametricRegion. -
(Merged) #19650: Added support to integrate over objects of geometry module.
-
(Merged) #19681: Added
ImplicitRegionclass to represent implicitly defined regions. -
(Megred) #19807: Implemented the algorithm for rational parametrization of conics.
-
(Merged) #20000: Added API of newly added classes to sympy’s documentation.
-
(Open) #19883: Allow
vector_integrateto handleImplicitRegionobjects. -
(Open) #20021: Add usage examples.
We had an important discussion regarding our initial approach and possible problems on issue #19320
Defining Regions with their parametric equations
The ParametricRegion class is used to represent a parametric region in space.
>>> from sympy.vector import CoordSys3D, ParametricRegion, vector_integrate
>>> from sympy.abc import r, theta, phi, t, x, y, z
>>> from sympy import pi, sin, cos, Eq
>>> C = CoordSys3D('C')
>>> circle = ParametricRegion((4*cos(theta), 4*sin(theta)), (theta, 0, 2*pi))
>>> disc = ParametricRegion((r*cos(theta), r*sin(theta)), (r, 0, 4), (theta, 0, 2*pi))
>>> box = ParametricRegion((x, y, z), (x, 0, 1), (y, -1, 1), (z, 0, 2))
>>> cone = ParametricRegion((r*cos(theta), r*sin(theta), r), (theta, 0, 2*pi), (r, 0, 3))
>>> vector_integrate(1, circle)
8*pi
>>> vector_integrate(C.x*C.x - 8*C.y*C.x, disc)
64*pi
>>> vector_integrate(C.i + C.j, box)
(Integral(1, (x, 0, 1), (y, -1, 1), (z, 0, 2)))*C.i + (Integral(1, (x, 0, 1), (y, -1, 1), (z, 0, 2)))*C.j
>>> _.doit()
4*C.i + 4*C.j
>>> vector_integrate(C.x*C.i - C.z*C.j, cone)
-9*pi
Integration over objects of geometry module
Many regions like circle are commonly encountered in problems of vector calculus. the geometry module of SymPy already had classes for some of these regions. The functionparametric_region_list was added to determine the parametric representation of such classes. vector_integrate can directly integrate objects of Point, Curve, Ellipse, Segment and Polygon class of geometry module.
>>> from sympy.geometry import Point, Segment, Polygon, Segment
>>> s = Segment(Point(4, -1, 9), Point(1, 5, 7))
>>> triangle = Polygon((0, 0), (1, 0), (1, 1))
>>> circle2 = Circle(Point(-2, 3), 6)
>>> vector_integrate(-6, s)
-42
>>> vector_integrate(C.x*C.y, circle2)
-72*pi
>>> vector_integrate(C.y*C.z*C.i + C.x*C.x*C.k, triangle)
-C.z/2
Implictly defined Regions
In many cases, it is difficult to determine the parametric representation of a region. Instead, the region is defined using its implicit equation. To represent implicitly defined regions, we implemented the ImplicitRegion class.
But to integrate over a region, we need to determine its parametric representation. the rationl_parametrization method was added to ImplicitRegion.
>>> from sympy.vector import ImplicitRegion
>>> circle3 = ImplicitRegion((x, y), (x-4)**2 + (y+3)**2 - 16)
>>> parabola = ImplicitRegion((x, y), (y - 1)**2 - 4*(x + 6))
>>> ellipse = ImplicitRegion((x, y), (x**2/4 + y**2/16 - 1))
>>> rect_hyperbola = ImplicitRegion((x, y), x*y - 1)
>>> sphere = ImplicitRegion((x, y, z), Eq(x**2 + y**2 + z**2, 2*x))
>>> circle3.rational_parametrization()
(8*t/(t**2 + 1) + 4, 8*t**2/(t**2 + 1) - 7)
>>> parabola.rational_parametrization()
(-6 + 4/t**2, 1 + 4/t)
>>> rect_hyperbola.rational_parametrization(t)
(-1 + (t + 1)/t, t)
>>> ellipse.rational_parametrization()
(t/(2*(t**2/16 + 1/4)), t**2/(2*(t**2/16 + 1/4)) - 4)
>>> sphere.rational_parametrization()
(2/(s**2 + t**2 + 1), 2*s/(s**2 + t**2 + 1), 2*t/(s**2 + t**2 + 1))
After PR #19883 gets merged, vector_integrate can directly work upon ImplicitRegion objects.
Future Work
-
In vector calculus, many regions cannot be defined using a single implicit equation. Instead, inequality or a combination of the implicit equation and conditional equations is required. For Example, a disc can only be represented using an inequality
ImplicitRegion(x**2 + y**2 < 9)and a semicircle can be represented asImplicitRegion(x**2 + y**2 -4) & (y < 0)). Support for such implicit regions needs to be added. -
The parametrization obtained using the
rational_parametrizationmethod in most cases is a large expression. SymPy’sIntegralis unable to work over such expressions. It takes too long and often does not returns a result. We need to either simplify the parametric equations obtained or fix Integral to handle them. -
Adding the support to plot objects of
ParamRegionandImplicitRegion
Conclusion
This summer has been a great learning experience. Writing a functionality, testing, debugging it has given me good experience in test-driven development. I think I realized many of the goals that I described in my proposal, though there were some major differences in the plan and actual implementation. Personally, I hoped I could have done better. I could not work much on the project during the last phase due to the start of the academic year.
I would like to thank Francesco for his valuable suggestions and being readily available for discussions. He was always very friendly and helpful. I would love to work more with him. S.Y Lee also provided useful suggestions.
Special thanks to Johann Mitteramskogler, Professor Sonia Perez Diaz, and Professor Sonia L.Rueda. They personally helped me in finding the algorithm for determining the rational parametrization of conics.
SymPy is an amazing project with a great community. I would mostly stick around after the GSoC period as well and continue contributing to SymPy, hopefully, exploring the other modules as well.
Comments are closed
August 29, 2020
00:00, Saturday, 29 August
This report summarises the work I have done during GSoC 2020 for SymPy. The links to the PRs are in chronological order. For following the progress made during GSoC, see my blog for the weekly posts.
About Me
I am Sachin Agarwal, a third-year undergraduate student majoring in Computer Science and Engineering from the Indian Institute of Information Technology, Guwahati.
Project Synopsis
My project involved fixing and amending functions related to series expansions and limit evaluation. For further information, my proposal for the project can be referred.
Pull Requests
This section describes the actual work done during the coding period in terms of merged PRs.
Phase 1
-
#19292 : This PR added a condition to
limitinfmethod ofgruntz.pyresolving incorrect limit evaluations. -
#19297 : This PR replaced
xreplace()withsubs()inrewritemethod ofgruntz.pyresolving incorrect limit evaluations. -
#19369 : This PR fixed
_eval_nseriesmethod ofmul.py. -
#19432 : This PR added a functionality to the
doitmethod oflimits.pywhich usesis_meromorphicfor limit evaluations. -
#19461 : This PR corrected the
_eval_as_leading_termmethod oftanandsecfunctions. -
#19508 : This PR fixed
_eval_nseriesmethod ofpower.py. -
#19515 : This PR added
_eval_rewrite_as_factorialand_eval_rewrite_as_gammamethods toclass subfactorial.
Phase 2
-
#19555 : This PR added
cdirparameter to handleseries expansionsonbranch cuts. -
#19646 : This PR rectified the
mrvmethod ofgruntz.pyandcancelmethod ofpolytools.pyresolvingRecursionErrorandTimeoutin limit evaluations. -
#19680 : This PR added
is_Powheuristic tolimits.pyto improve the limit evaluations ofPow objects. -
#19697 : This PR rectified
_eval_rewrite_as_tractablemethod ofclass erf. -
#19716 : This PR added
_singularitiestoLambertWfunction. -
#18696 : This PR fixed errors in assumptions when rewriting
RisingFactorial/FallingFactorialasgammaorfactorial.
Phase 3
-
#19741 : This PR reduced
symbolic multiples of piintrigonometric functions. -
#19916 : This PR added
_eval_nseriesmethod tosinandcos. -
#19963 : This PR added
_eval_is_meromorphicmethod to theclass BesselBase. -
#18656 : This PR added
Raabe's Testto theconcretemodule. -
#19990 : This PR added
_eval_is_meromorphicand_eval_aseriesmethods toclass lowergamma,_eval_is_meromorphicand_eval_rewrite_as_tractablemethods toclass uppergammaand rectified theevalmethod ofclass besselk. -
#20002 : This PR fixed
_eval_nseriesmethod oflog.
Miscellaneous Work
This section contains some of my PRs related to miscellaneous issues.
-
#19447 : This PR added some required testcases to
test_limits.py. -
#19537 : This PR fixed a minor
performanceissue. -
#19604 : This PR fixed
AttributeErrorin limit evaluation.
Reviewed Work
This section contains some of the PRs which were reviewed by me.
Issues Opened
This section contains some of the issues which were opened by me.
- #19670 :
Poly(E**100000000)is slow to create. - #19752 :
gammasimpcan be improved forintegervariables.
Examples
This section describes the bugs fixed and the new features added during GSoC.
Fixed Limit Evaluations
>>> from sympy import limit, limit_seq
>>> n = Symbol('n', positive=True, integer=True)
>>> limit(factorial(n + 1)**(1/(n + 1)) - factorial(n)**(1/n), n, oo)
exp(-1) # Previously produced 0
>>> n = Symbol('n', positive=True, integer=True)
>>> limit(factorial(n)/sqrt(n)*(exp(1)/n)**n, n, oo)
sqrt(2)*sqrt(pi) # Previously produced 0
>>> n = Symbol('n', positive=True, integer=True)
>>> limit(n/(factorial(n)**(1/n)), n, oo)
exp(1) # Previously produced oo
>>> limit(log(exp(3*x) + x)/log(exp(x) + x**100), x, oo)
3 # Previously produced 9
>>> limit((2*exp(3*x)/(exp(2*x) + 1))**(1/x), x, oo)
exp(1) # Previously produced exp(7/3)
>>> limit(sin(x)**15, x, 0, '-')
0 # Previously it hanged
>>> limit(1/x, x, 0, dir="+-")
zoo # Previously raised ValueError
>>> limit(gamma(x)/(gamma(x - 1)*gamma(x + 2)), x, 0)
-1 # Previously it was returned unevaluated
>>> e = (x/2) * (-2*x**3 - 2*(x**3 - 1) * x**2 * digamma(x**3 + 1) + 2*(x**3 - 1) * x**2 * digamma(x**3 + x + 1) + x + 3)
>>> limit(e, x, oo)
1/3 # Previously produced 5/6
>>> a, b, c, x = symbols('a b c x', positive=True)
>>> limit((a + 1)*x - sqrt((a + 1)**2*x**2 + b*x + c), x, oo)
-b/(2*a + 2) # Previously produced nan
>>> limit_seq(subfactorial(n)/factorial(n), n)
1/e # Previously produced 0
>>> limit(x**(2**x*3**(-x)), x, oo)
1 # Previously raised AttributeError
>>> limit(n**(Rational(1, 1e9) - 1), n, oo)
0 # Previously it hanged
>>> limit((1/(log(x)**log(x)))**(1/x), x, oo)
1 # Previously raised RecursionError
>>> e = (2**x*(2 + 2**(-x)*(-2*2**x + x + 2))/(x + 1))**(x + 1)
>>> limit(e, x, oo)
exp(1) # Previously raised RecursionError
>>> e = (log(x, 2)**7 + 10*x*factorial(x) + 5**x) / (factorial(x + 1) + 3*factorial(x) + 10**x)
>>> limit(e, x, oo)
10 # Previously raised RecursionError
>>> limit((x**2000 - (x + 1)**2000) / x**1999, x, oo)
-2000 # Previously it hanged
>>> limit(((x**(x + 1) + (x + 1)**x) / x**(x + 1))**x, x, oo)
exp(exp(1)) # Previously raised RecursionError
>>> limit(Abs(log(x)/x**3), x, oo)
0 # Previously it was returned unevaluted
>>> limit(x*(Abs(log(x)/x**3)/Abs(log(x + 1)/(x + 1)**3) - 1), x, oo)
3 # Previously raised RecursionError
>>> limit((1 - S(1)/2*x)**(3*x), x, oo)
zoo # Previously produced 0
>>> d, t = symbols('d t', positive=True)
>>> limit(erf(1 - t/d), t, oo)
-1 # Previously produced 1
>>> s, x = symbols('s x', real=True)
>>> limit(erf(s*x)/erf(s), s, 0)
x # Previously produced 1
>>> limit(erfc(log(1/x)), x, oo)
2 # Previously produced 0
>>> limit(erf(sqrt(x)-x), x, oo)
-1 # Previously produced 1
>>> a, b = symbols('a b', positive=True)
>>> limit(LambertW(a), a, b)
LambertW(b) # Previously produced b
>>> limit(uppergamma(n, 1) / gamma(n), n, oo)
1 # Previously produced 0
>>> limit(besselk(0, x), x, oo)
0 # Previously produced besselk(0, oo)
Rewrote Mul._eval_nseries()
>>> e = (exp(x) - 1)/x
>>> e.nseries(x, 0, 3)
1 + x/2 + x**2/6 + O(x**3) # Previously produced 1 + x/2 + O(x**2, x)
>>> e = (2/x + 3/x**2)/(1/x + 1/x**2)
>>> e.nseries(x, n=3)
3 - x + x**2 + O(x**3) # Previously produced 3 + O(x)
Rewrote Pow._eval_nseries()
>>> e = (x**2 + x + 1) / (x**3 + x**2)
>>> series(e, x, oo)
x**(-5) - 1/x**4 + x**(-3) + 1/x + O(x**(-6), (x, oo))
# Previously produced x**(-5) + x**(-3) + 1/x + O(x**(-6), (x, oo))
>>> e = (1 - 1/(x/2 - 1/(2*x))**4)**(S(1)/8)
>>> e.series(x, 0, n=17)
1 - 2*x**4 - 8*x**6 - 34*x**8 - 152*x**10 - 714*x**12 - 3472*x**14 - 17318*x**16 + O(x**17)
# Previously produced 1 - 2*x**4 - 8*x**6 - 34*x**8 - 24*x**10 + 118*x**12 - 672*x**14 - 686*x**16 + O(x**17)
Added Series Expansions and Limit evaluations on Branch-Cuts
>>> asin(I*x + I*x**3 + 2)._eval_nseries(x, 3, None, 1)
-asin(2) + pi - sqrt(3)*x/3 + sqrt(3)*I*x**2/9 + O(x**3)
>>> limit(log(I*x - 1), x, 0, '-')
-I*pi
Rectified ff._eval_rewrite_as_gamma() and rf._eval_rewrite_as_gamma()
>>> n = symbols('n', integer=True)
>>> combsimp(RisingFactorial(-10, n))
3628800*(-1)**n/factorial(10 - n) # Previously produced 0
>>> ff(5, y).rewrite(gamma)
120/Gamma(6 - y) # Previously produced 0
Added Raabe’s Test
>>> Sum(factorial(n)/factorial(n + 2), (n, 1, oo)).is_convergent()
True # Previously raised NotImplementedError
>>> Sum((-n + (n**3 + 1)**(S(1)/3))/log(n), (n, 1, oo)).is_convergent()
True # Previously raised NotImplementedError
Rewrote log._eval_nseries()
>>> f = log(x/(1 - x))
>>> f.series(x, 0.491, n=1).removeO()
-0.0360038887560022 # Previously raised ValueError
Future Work
- Refactoring high level functions like
series,nseries,lseriesandaseries. - Add
_eval_is_meromorphicmethod or_singularitiesto as many special functions as possible. - Work can be done to resolve the issues opened by me (listed above).
Conclusion
This summer has been a great learning experience and has helped me get a good exposure of test-driven development. I plan to continue contributing to SymPy and will also try to help the new contributors. I am grateful to my mentors, Kalevi Suominen and Sartaj Singh for reviewing my work, giving me valuable suggestions, and being readily available for discussions.
August 28, 2020
00:00, Friday, 28 August
Key highlights of this week’s work are:
-
- This was a long pending issue. It was necessary to refactor the
_eval_nseriesmethod oflogto fix some minor issues.
- This was a long pending issue. It was necessary to refactor the
This marks the end of Phase-3 of the program.
Finally, this brings us to the end of Google Summer of Code and I am really thankful to the SymPy Community and my mentor Kalevi Suominen for always helping and supporting me.
August 21, 2020
00:00, Friday, 21 August
Key highlights of this week’s work are:
-
- This was a long pending issue.
Raabe's Testhelps to determine theconvergenceof a series. It has been added to theconcretemodule to handle those cases when theratio testbecomesinconclusive.
- This was a long pending issue.
-
Fixed limit evaluations related to lowergamma, uppergamma and besselk function
-
We added
_eval_is_meromorphicmethod to classlowergammaanduppergammaso that some of thelimitsinvolvinglowergammaanduppergammafunctions get evaluated using themeromorphic checkalready present in the limit codebase. Now, to makelowergammaanduppergammafunctionstractablefor limit evaluations,_eval_aseriesmethod was added tolowergammaand_eval_rewrite_as_tractabletouppergamma.Finally, we also
rectifiedtheevalmethod of classbesselk, so thatbesselk(nu, oo)automatically evaluates to0.
-
August 14, 2020
00:00, Friday, 14 August
Key highlights of this week’s work are:
-
Added _eval_is_meromorphic() to bessel function
- We added the
_eval_is_meromorphicmethod to the classBesselBaseso that some of the limits involvingbessel functionsget evaluated using themeromorphic checkalready present in the limit codebase.
- We added the
August 07, 2020
00:00, Friday, 07 August
Key highlights of this week’s work are:
-
Fixed periodicity of trigonometric function
- This was a long pending issue. The
_peeloff_pimethod ofsinhad to be rectified to reduce the symbolic multiples of pi in trigonometric functions. As a result of this fix, nowsin(2*n*pi + 4)automatically evaluates tosin(4), whennis aninteger.
- This was a long pending issue. The
-
Fixed limit evaluations related to trigonometric functions
- In this PR, we added
_eval_nseriesmethod to bothsinandcosto make the limit evaluations more robust when it comes to trigonometric functions. We also added a piece of code tocos.evalmethod so that thelimit of cos(m*x), wheremisnon-zero, andxtends toooevaluates toAccumBounds(-1, 1).
- In this PR, we added
July 13, 2020
00:00, Monday, 13 July
This week, I spent most of my time reading about algorithms to parametrize algebraic curves and surfaces.
Parametrization of algebraic curves and surfaces
In many cases, it is easiar to define a region using an implicit equation over its parametric form. For example, a sphere can be defined with the implict eqution x2 + y2 + z2 = 4. Its parametric equation although easy to determine are tedious to write.
To integrate over implicitly defined regions, it is necessary to determine its parametric representation. I found the report on conversion methods between parametric and implicit curves from Christopher M. Hoffmann very useful. It lists several algorithms for curves and surfaces of different nature.
Every plane parametric curve can be expressed as an implicit curve. Some, but not all implicit curves can be expressed as parametric curves. Similarly, we can state of algebraic surfaces. Every plane parametric surface can be expressed as an implicit surface. Some, but not all implicit surfaces can be expressed as parametric surfaces.
Conic sections
One of the algorithm to parametrize conics is given below:
- Fix a point p on the conic. Consider the pencil of lines through p. Formulate the line equations.
- Substitute for y in the conic equation, solve for x(t).
- Use the fine equations to determine y(t).
The difficult part is to find a point on the conic. For the first implementation, I just iterated over a few points and selected the point which satisfied the equation. I need to implement an algorithm that can find a point on the curve.
The parametric equations determined by the algoithm are large and complex expressions. The vector_integrate function in most cases is taking too much time to calculate the integral. I tried fixing this using simplification functions in SymPy like trigsimp and expand_trig. The problem still persist.
>>> ellipse = ImplicitRegion(x**2/4 + y**2/16 - 1, x, y)
>>> parametric_region_list(ellipse)
[ParametricRegion((8*tan(theta)/(tan(theta)**2 + 4), -4 + 8*tan(theta)**2/(tan(theta)**2 + 4)), (theta, 0, pi))]
>>> vector_integrate(2, ellipse)
### It gets stuck
The algorithm does not work for curves of higher algebraic degree like cubics.
Monoids
A monoid is an algebraic curve of degree n that has a point of multiplicity n -1. All conics and singular cubics are monoids. Parametrizing a monoid is easy if the special point is known.
To parametrize a monoid, we need to determine a singular point of n - 1 multiplicity. The singular points are those points on the curve where both partial derivatives vanish. A singular point (xo, yo) of a Curve C is said to be of multiplicity n if all the partial derivatives off to order n - 1 vanish there. This paper describes an algorithm to calculate such a point but the algorithm is not trivial.
Next week’s goal
My goal is to complete the work on parametrizing conics. If I can find a simple algorithm to determine the singular point of the required multiplicity, I will start working on it.
Comments are closed
00:00, Monday, 13 July
My goal for this week was to add support of integration over objects of geometry module.
Integrating over objects of geometry module
In my GSoC proposal, I mentioned implementing classes to represent commonly used regions. It will allow a user to easily define regions without bothering about its parametric representation. SymPy’s geometry module has classes to represent commonly used geometric entities like line, circle, and parabola. Francesco told me it would be better to add support to use these classes instead. The integral takes place over the boundary of the geometric entity, not over the area or volume enclosed.
My first approach was to add a function parametric_region to the classes of geometry module. Francesco suggested not to modify the geometry module as this would make it dependent on vector module. We decided to implement a function parametric_region to return a ParametricRegion of objects of geometry module.
I learned about the singledispatch decorater of python. It is used to create overloaded functions.
Polygons cannot be represented as a single ParametricRegion object. Each side of a polygon has its separate parametric representation. To resolve this, we decided to return a list of ParametricRegion objects.
My next step was to modify vector_integrate to support objects of geometry module. This was easy and involved calling the parametric_region_list function and integrating each ParametricRegion object.
The PR for the work has been merged. Note that we still do not have any way to create regions like disc and cone without their parametric representation. To support such regions, I think we need to add new classes to the geometry module.
Next week’s goal
I plan to work on implementing a class to represent implicitly defined regions.
Comments are closed
00:00, Monday, 13 July
The first phase of GSoC is over. We can now integrate the scalar or vector field over a parametrically defined region.
The PR for ParametricIntegral class has been merged. In my last post, I mentioned about a weird error. It turns out that I was importing pi as a symbol instead of as a number. Due to this, the expressions were returned unevaluated causing tests to fail.
The route ahead
I had a detailed discussion with Francesco about the route ahead. My plan has drifted from my proposal because we have decided not to implement new classes for special regions like circles and spheres. The geometry module already has classes to represent some of these regions. We have decided to use these classes.
Francesco reminded me that we have to complete the documentation of these changes. I was planning to complete the documentation in the last phase. But I will try completing it soon as I may forget some details.
We also discussed adding support to perform integral transformation using Stoke’s and Green’s theorem. In many cases, such transformation can be useful but adding them may be outside the scope of this project.
I have convinced Francesco on adding classes to represent implicitly defined regions. It might be the most difficult part of the project but hopefully, it will be useful to SymPy users.
Next week’s goal
My next week’s goal is to make a PR for adding suppor to integrate over objects of the geometry module.
Comments are closed
July 12, 2020
00:00, Sunday, 12 July
Key highlights of this week’s work are:
-
Fixed incorrect limit evaluation related to LambertW function
- This was a minor bug fix. We added the
_singularitiesfeature to theLambertWfunction so that its limit gets evaluated using themeromorphic checkalready present in the limit codebase.
- This was a minor bug fix. We added the
-
Fixed errors in assumptions when rewriting RisingFactorial / FallingFactorial as gamma or factorial
- This was a long pending issue. The rewrite to
gammaorfactorialmethods ofRisingFactorialandFallingFactorialdid not handle all the possible cases, which caused errors in some evaluations. Thus, we decided to come up with a proper rewrite usingPiecewisewhich accordingly returned the correct rewrite depending on the assumptions on the variables. Handling such rewrites usingPiecewiseis never easy, and thus there were a lot of failing testcases. After spending a lot of time debugging and fixing each failing testcase, we were finally able to merge this.
- This was a long pending issue. The rewrite to
This marks the end of Phase-2 of the program. I learnt a lot during these two months and gained many important things from my mentors.